Introduction
Proteomics, the large-scale study of proteomes, is a highly valuable tool in biomedical research, but because most proteomics approaches are based on mass spectrometry (MS), the analytical chemistry nature can be a barrier to some biologists and physician-scientists. In this series of three blog articles, I would like to introduce MS-based proteomics data by providing an overview of the associated proteomics data repository landscape. I’ll also provide some guidance and tips for navigating the landscape for researchers looking for MS-based proteomics data and/or generating MS-based proteomics data and needing or wanting to share it.
In the first blog article, I am going to introduce the ProteomeXchange (PX) Consortium and its most important member – the PRIDE Database. In the second blog article, I will cover the rest of the repositories in the PX Consortium. In the third and final blog article, I will highlight additional repositories outside of the PX Consortium that are accepting proteomics data and their role in the proteomics data sharing landscape.
ProteomeXchange Consortium
Since 2012, the PX Consortium has become the international resource for standardizing data submission and dissemination of public MS proteomics data. Currently, the consortium consists of six members (PRIDE, PeptideAtlas/PASSEL, MassIVE, iPOST, iPROX and Panorama Public), and a common portal called ProteomeCentral provides search capabilities for public datasets in all member repositories. As of November 2024, there are more than 38,400 datasets publicly available in ProteomeCentral (Fig. 1).
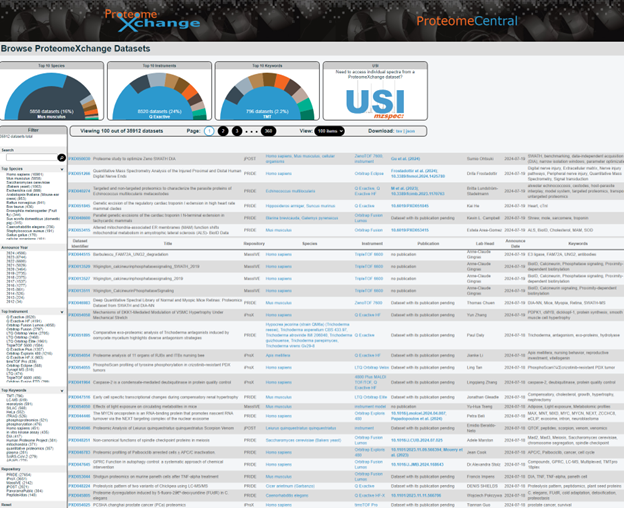
Each original dataset in ProteomeCentral is assigned a PX Accession Number beginning with “PXD” followed by six digits (Fig. 2), while datasets from reanalysis projects are assigned accession numbers beginning with “PRXD”. In addition to the accession number, each dataset record contains a wealth of metadata, including a dataset summary, dataset history, publication list, keyword list, contact list, full dataset link list and a repository record list. Users can also download all of the metadata as an XML file.
When searching ProteomeCentral, the search results can be filtered by species, announce year, instrument, keywords, and repository, and search results can be downloaded as a TSV or JSON file.
Many mass spectra in ProteomeCentral are also assigned a Universal Spectrum Identifier (USI). Users can use the USI portal (Fig. 1, top right) to inspect observed mass spectra archived in ProteomeCentral or the six consortium members, as well as predicted mass spectra generated from MS2PIP and Seq2MS. The format of the USI is mzspec:{PX Accession}:{MS Run}:scan:{Scan Number}:{Peptidoform}/{Charge}, and the observed and predicted spectra for mzspec:PXD000561:Adult_Frontalcortex_bRP_Elite_85_f09:scan:17555:VLHPLEGAVVIIFK/2 are shown in Fig. 3 and Fig. 4, respectively.

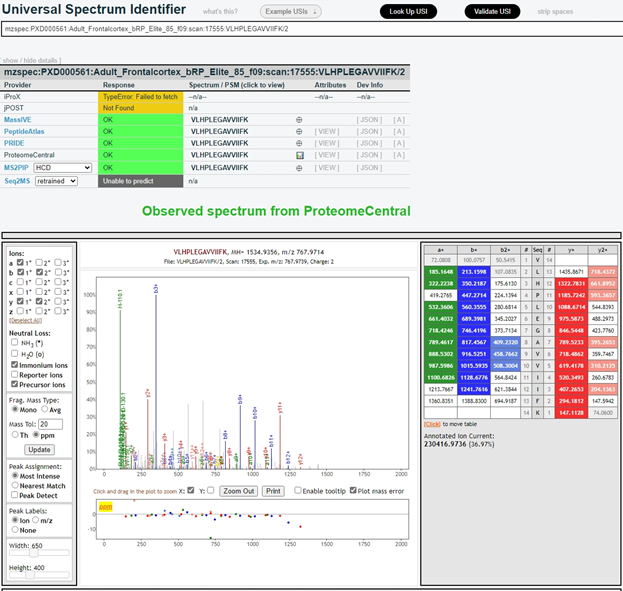
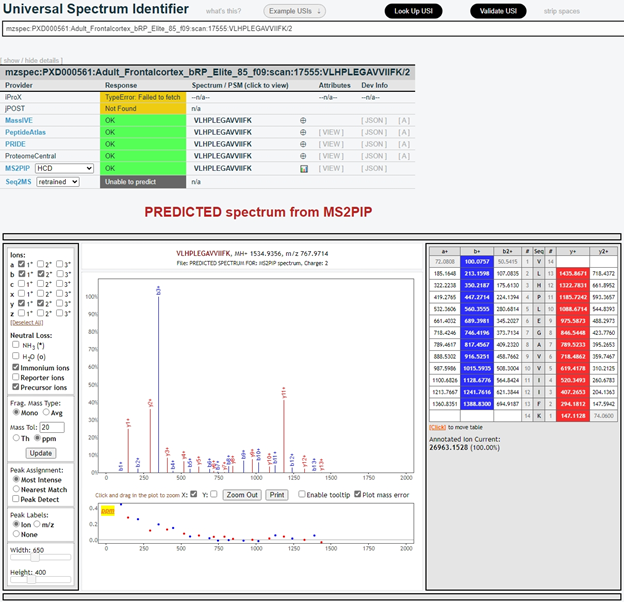
In the next section, I am going to provide an overview of the PRIDE Database, the most widely used data repository in the PX Consortium.
The PRoteomics IDEntifications (PRIDE) Database
The PRIDE Database was established by the European Bioinformatics Institute (EMBL-EBI) in 2004. Since then, it has become the most widely used proteomics data repository, with about 77% of the datasets in ProteomeCentral coming from PRIDE. PRIDE stores datasets coming from all MS-based proteomics experimental approaches, with a focus on discovery-driven techniques such as data dependent acquisition (DDA), data independent acquisition (DIA), and bottom-up proteomics, but additional approaches like top-down proteomics and MS imaging are included as well. Users can search for PRIDE datasets in either the PRIDE Archive Portal or in ProteomeCentral.
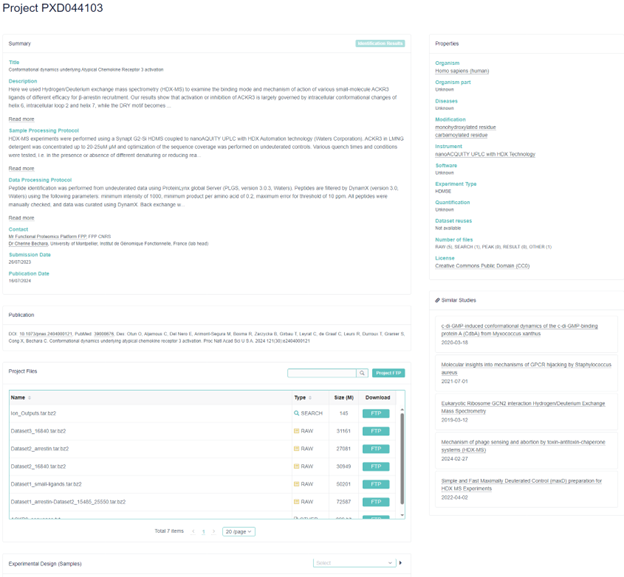
The PRIDE Database captures more dataset metadata than ProteomeCentral (Fig. 5), including sample processing protocol, data processing protocol, organism part, diseases, software, experiment type, quantification and dataset reuses. Submitters can also label datasets in the PRIDE Database with one or more project tags, with “Biological”, “Biomedical”, “Technical” and “Human” being the most commonly used tags.
The PRIDE Archive Portal also offers some additional search fields that are not present in ProteomeCentral, such as organism part, diseases, modification, country, and submission type. The project tag values can be searched as well. Search results in the PRIDE Archive Portal can also be sorted by accession number, title, relevance, submission date, or publication date. Unlike in ProteomeCentral, search results in the PRIDE Archive Portal can only be downloaded as JSON files.
All PRIDE datasets are shared under the Creative Commons CC0 Public Domain Dedication or the EBI terms of use. The PRIDE Database also automatically provides links to five studies that are similar to a given dataset (Fig. 5). Like ProteomeCentral, the PRIDE Database also has a USI portal for spectrum searches. The PRIDE Spectrum Libraries contain the consensus spectra generated from the public experiments in the PRIDE Archive Portal. However, these libraries have not been updated since 2015. In addition, the PRIDE Crosslinking Database allows users to visualize a small set of cross-linking MS data.
In the next blog article, I am going to cover the other repositories in the PX Consortium. If you have questions regarding sharing and re-using proteomics data, please do not hesitate to contact the Data Management and Sharing team at Becker Library at BeckerDMS@wustl.edu.
