In the first two parts of this blog series, I introduced the main player in the proteomics data repository ecosystem, the ProteomeXchange (PX) Consortium. Outside of the PX Consortium, there are an additional handful of repositories holding proteomics data. These additional repositories serve special cases or certain scientific communities, but it may be useful for researchers to know that there are alternatives. I will also cover WashU Medicine’s institutional data repository, Digital Commons Data@Becker, as an option for School of Medicine researchers to share proteomics data, including sensitive proteomics data from human participants.
1. Proteomic Data Commons
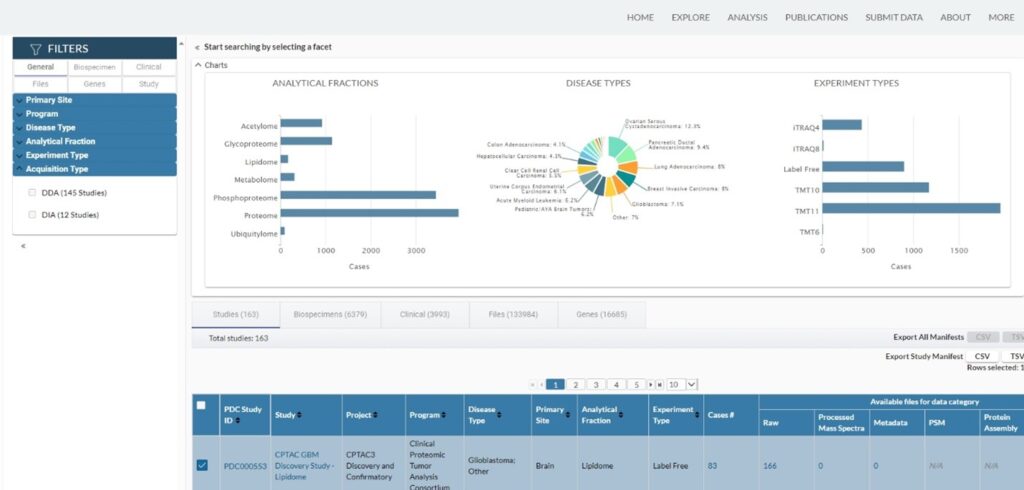
Launched in 2020, Proteomic Data Commons (PDC) is one of several repositories within the National Cancer Institute’s (NCI) Cancer Research Data Commons (CRDC), a secure cloud-based infrastructure featuring diverse datasets and innovative analytic tools for cancer research. The CRDC enables researchers to link proteomics data with other datasets and to submit, collect, analyze, store, and share data throughout the cancer data ecosystem. Besides proteomics data, PDC also accepts metabolomics and lipidomics data. As of September 2024, there are more than 160 studies in PDC, most of which are contributed by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) (Figure 1).
PDC offers a few data analysis tools, including “Explore Quantitation Data”, “Common Data Analysis Pipeline (CDAP)”, and “cProSite”. A subset of CPTAC datasets can also be visualized in a browser for “Peptide Genome mapping”. In addition, a third-party tool named “PepQuery” serves as a universal targeted peptide search engine for many CPTAC datasets. These tools are still in the active development phase, so their functionality is likely to improve in the future. Researchers can also analyze PDC data in the NCI Cloud Resources.
PDC data submission and data use are governed by Creative Commons CC-BY 4.0 licensing terms. Researchers who are interested in submitting data to PDC need to first make a data submission request. Once the NCI approves the request, PDC will set up a “Program” for researchers to submit data. While most datasets submitted to PDC are generated with NCI funding, PDC will also consider cancer-related datasets generated by projects without NCI funding. I would recommend submitting proteomics data to PDC if related datasets are being submitted to other components of CRDC, such as Genomic Data Commons (GDC) and Imaging Data Commons (IDC).
2. AD Knowledge Portal and Synapse Platform
Some domain specific repositories also host proteomics data. For example, the AD Knowledge Portal supported by the National Institute on Aging (NIA) currently has 27 studies containing proteomics data. Prior approval is required for data submission.
The backend of the AD Knowledge Portal is the Synapse platform, created by Sage Bionetworks. Synapse serves as the backend for several other domain-specific data portals developed (Figure 2), and some portals are currently hosting or plan to host proteomics data. Interested researchers should contact the portals for details.

3. European Genome-phenome Archive (EGA)
Since the introduction of the General Data Protection Regulation (GDPR) by the European Union in 2016, there is an increasing awareness in the scientific community that proteomics data from human participants may potentially contain personal identifiable information (PII). As a result, researchers in the European Union have begun to deposit sensitive proteomics data from human participants into the European Genome-phenome Archive (EGA).
Launched by the European Bioinformatics Institute (EMBL-EBI) in 2008, EGA is a repository for long term secure archiving of all types of potentially identifiable genetic, phenotypic, and clinical data resulting from biomedical research. Acting as a data distributor, EGA allows data submitters to maintain control over who has access to data and under which conditions. As of today, EGA hosts over eight thousand studies, including a few proteomics and metabolomics studies.
If you are submitting proteomics data to EGA, be aware that there are at least two obstacles during the submission process. Firstly, the EGA Data Submission Portal currently only supports genomics data and does not utilize metadata standards for proteomics data. Secondly, EGA’s data access model is designed to fit GDPR and is not necessarily suitable for institutions in the United States. While submitting proteomics data to EGA is currently possible, I would only recommend doing so when proteomics data is generated from samples that are under GDPR.
4. Digital Commons Data@Becker
The three main scenarios for scientific data sharing requiring controlled or restricted access are: (1) data could potentially uniquely match to a single individual, thus pose a de-identification risk; (2) informed consent forms specify that the data would be under controlled access; (3) data were generated from samples subjected to regulations, such as GDPR.
Digital Commons Data@Becker, our School of Medicine institutional data repository managed by Becker Library, can accommodate datasets requiring restricted access, including proteomics datasets. If you would like to share potentially sensitive proteomics data from human participants, please contact our Becker Library Data Management and Sharing Team at BeckerDMS@wustl.edu. We will work with the Human Research Protection Office to determine what is appropriate for your dataset and provide guidance and curation assistance to ensure your data is shared according to the FAIR (Finable, Accessible, Interoperable, Reusable) data principles.
The following table summarizes the specifics of the repositories mentioned in this article.
| Name | Country | Access Type | Main Features |
|---|---|---|---|
| PRIDE Database | EU/UK | Open | Default repository for most data |
| iProX | China | Open | Suitable for Chinese nationals data |
| MassIVE | USA | Open | Suitable for experts and re-analysis |
| jPOST | Japan | Open | Accepting non-MS data |
| Panorama Public | USA | Open | Suitable for Skyline software data |
| PeptideAtlas/PASSEL | USA | Open | Suitable for SRM/MRM data |
| PDC | USA | Open | Suitable for cancer data |
| AD Knowledge Portal | USA | Controlled | Suitable for Alzheimer’s disease data |
| EGA | EU | Controlled | Possible for samples under GDPR |
| Digital Commons Data@Becker | USA | Controlled or Open | Suitable for samples requiring controlled access |
This concludes my blog series about proteomics data repositories. If you have questions related to sharing proteomics data, please do not hesitate to contact me (at jianx@wustl.edu) or our Becker Library Data Management and Sharing team (at BeckerDMS@wustl.edu).
