A plethora of learning resources for bioinformatics and genomics are available on the web, but did you know that you can find a wide variety of free learning resources on campus here at WashU? Do you need to learn fundamentals of bioinformatics and computational genomics? Are you interested in learning more about techniques and tools used in genomics research and data analysis? Would you like to connect with a WashU community of like-minded researchers on campus? This blog highlights a variety of sources of free bioinformatics educational content and learning opportunities made available on campus for WashU researchers.
Becker Library’s Research Computing service offers free genomic data analysis workshops which introduce learners to the R programming language, Bioconductor packages and includes hands-on demonstrations that enhance learning. Each workshop is described below:
- Introduction to R – Bioconductor Workshop: Introduces Bioconductor and demonstrates how to install, load, and manage Bioconductor packages as well as basic genomic data exploration with the VariantAnnotation package. Lecture materials and a recording of the workshop can be found here.
- Using R for Downstream Analysis of Single Cell Data Workshop: Led by Madhurima Kaushal, Bioinformatics Scientist at WashU’s Institute for Informatics, Data Science & Biostatistics (I2DB), this workshop introduced a series of steps for single cell data exploration – from cell clustering tools and techniques to cell annotations and pathway analysis to help determine cell types and cell markers. The workshop materials can be found here.
- Single Cell RNAseq Data Analysis Using R Workshop: Led by Madhurima Kaushal, Bioinformatics Scientist at WashU’s Institute for Informatics, Data Science & Biostatistics (I2DB), the session focused on using R for scRNAseq data analysis and included live demonstrations on how to analyze and interpret scRNASeq data using the R package Seurat (Version 5). The workshop recording and additional materials are located here.
- RNA-seq Data Analysis Using Bioconductor Workshop: Led by Madhurima Kaushal, Bioinformatics Scientist at WashU’s Institute for Informatics, Data Science & Biostatistics (I2DB), the training covered how to use the Bioconductor EdgeR and ShortRead packages for RNA sequencing data analysis. The hands-on demonstrations included various data formats, data input and output, data analysis, and visualization. The workshop materials are located here.
- Single-Cell RNA-Sequencing (scRNA-Seq) Data Analysis Using R and RIS Compute Platform Workshop: Led by Madhurima Kaushal, Bioinformatics Scientist at WashU’s Institute for Informatics, Data Science & Biostatistics (I2DB), this workshop discussed analyses of scRNA-Seq data using R on the RIS Scientific Compute Platform. The session also covered how to create and use a Docker container image for scRNA-Seq data analysis to submit a job that sets up RStudio on the RIS Scientific Compute Platform. Finally, R packages were used to explore and visualize the data. All the workshop materials are here.
- Introduction to 16S Amplicon Sequence Variant Analysis using R Workshop: Presented by Dr. Brigida Rusconi, an Instructor in the Department of Pediatrics at Washington University School of Medicine, this workshop introduced common 16S metrics and good practices for 16S data analysis. The session included hands-on demonstrations on how to use RStudio docker images with pre-installed packages and scripts to analyze a mock 16S V4 data set. All workshop materials and a recording are found here.
Bioinformatics Workshop Series (BFX) is an on-campus, year-long bioinformatics training course that focuses on teaching the practical skills that scientists need to analyze their own genomic data. The focus is on giving users hands on experience with software, data formats, and tools for visualization that will help them turn data into insight. It is open to anyone, and participants are welcome to join throughout the year. Recorded lectures and assignments are stored on the course website for later access.
The BFX is sponsored by the Institute of Clinical and Translational Sciences (ICTS) and it is one of the ICTS Precision Health educational offerings along with the Genomics in Medicine Seminar series and Precision Health Pathway. Each BFX lecture is recorded, and all weekly lecture materials are stored on the BFX course github page. Shown below is a picture of the agenda for the BFX Week 1 lecture which is an Intro to Bioinformatics and Computer Setup, to get learners prepared for the subsequent hands-on heavy lecture weeks that follow (Figure 1).
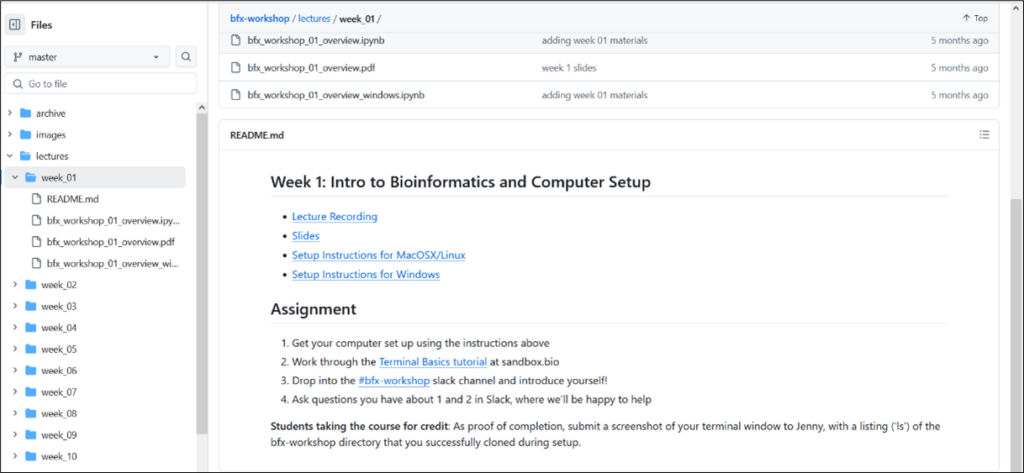
Interested participants can join the BFX course anytime throughout the year and support can be provided to help learners get caught up if needed.
The ICTS Precision Health Slack is a community where new and experienced users can converse, get and share knowledge, and connect with other researchers across campus. There are channels for high performance computing (#compute), data sharing, and specific technologies like #r, #python, or #docker. Everyone at WashU is welcome, and people can join with their @wustl.edu address (not a personal email) using this url – https://join.slack.com/t/ictsprecisionhealth/shared_invite/zt-1kj8067zp-hVStcLTAiR5yXNT1Bu_vDw.
If you’d like to have access to the ICTS Precision Health Slack, please email Jenny McKenzie at j.mckenzie@wustl.edu.
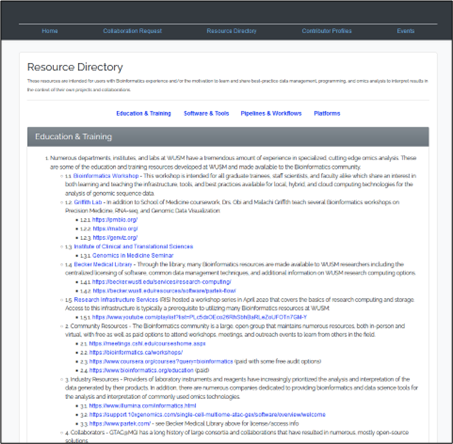
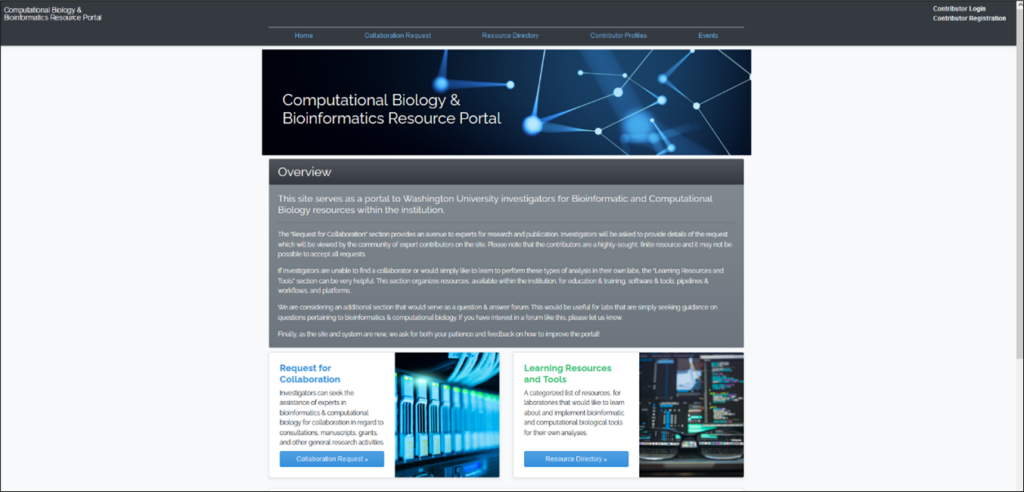
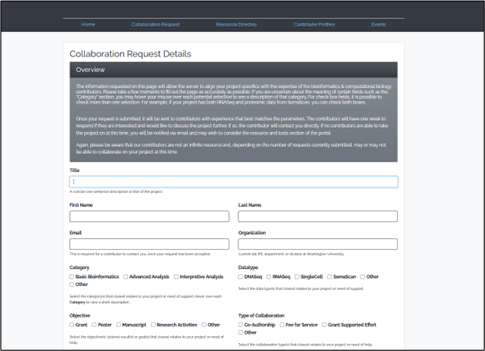
Computational Biology and Bioinformatics Resource Portal is a directory for bioinformatics analysis and collaboration created by WashU researchers for WashU researchers (to view this site, you must be connected to the WashU network or VPN). See Figures 2, 3 and 4.
WashU researchers can access the Resource Directory (to view this site, you must be connected to the WashU network or VPN) section of the portal which has learning resources, software, tools, pipelines, workflows, and platforms. These resources are intended for users with bioinformatics experience and users who are motivated to learn and share best-practice management, programming and omics data analysis to interpret results in the context of their projects and collaborations.
WashU researchers can use the portal to request collaborations if they are interested in collaborating with other researchers. Researchers need to first review the collaboration request details before completing the collaboration request form (to view this site, you must be connected to the WashU network or VPN).
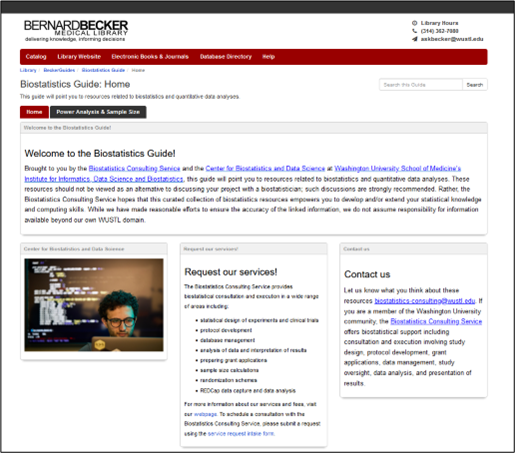
Biostatistics Guide provides resources and references for frequently asked questions related to biostatistical analyses for research projects including genomics projects (Figure 5). The Biostatistics Consulting Service within the Center for Biostatistics and Data Science (CBDS) created this guide in collaboration with Becker Library and hopes that it will empower researchers on campus who are interested in developing and/or extending their statistical knowledge and computing skills with self-directed online learning resources. Learn more here.
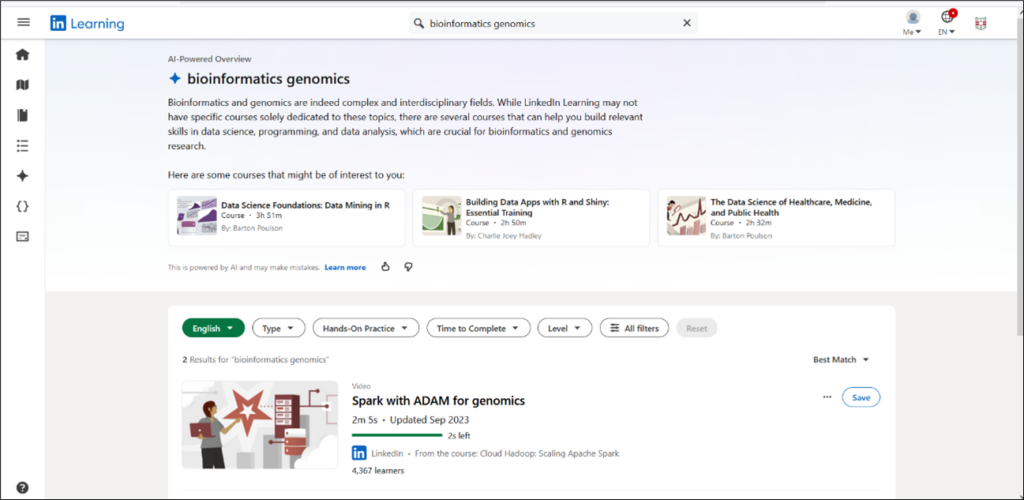
LinkedIn Learning is available to all members of the WashU community. While at this time LinkedIn Learning does not have a lot of content on genomics, it’s video tutorials can help you explore related areas crucial to genomics research (Figure 6). Users can perform a word search to find video tutorials on a variety of topics including bioinformatics, genomics and topics related to genomics research such as data science, data analysis, programming, and data visualization.
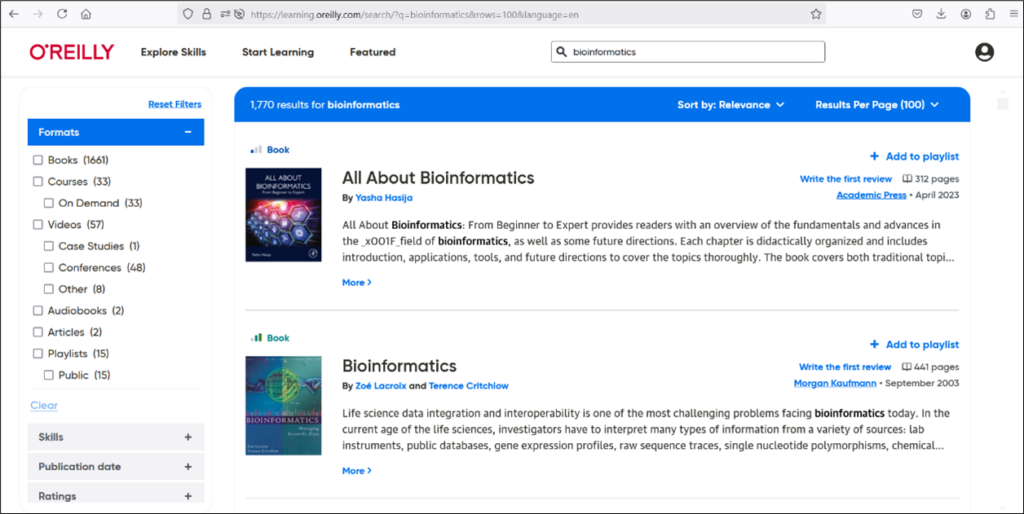
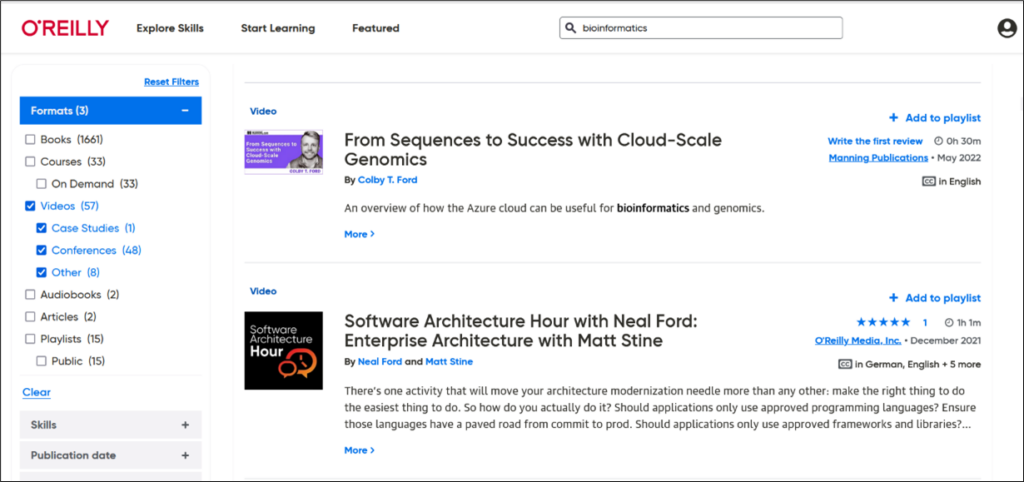
O’Reilly Online Learning Academic Library Edition is a collection containing over 30,000 full-text e-books and videos on computing, databases, networking, operating systems, programming, software engineering, .NET, Cisco products, Web design, Java, XML and more. It is a fully searchable platform where you can search for videos, books, and even snippets of code. You can access the O’Reilly/Safari Tech Books Online by using a valid Washington University email and create a free user account to retain notes etc., in the e-books you have used. For offline reading, you can use the O’Reilly Online Learning mobile app. More information about the mobile app is here: https://www.oreilly.com/online-learning/apps.html.
Follow the steps outlined in this blog to search for learning materials for a variety of skills and topics. You can search for e-books and videos on bioinformatics, computational genomics or content that can help you explore areas related to genomics such as data analysis, programming and data visualization (Figures 7 and 8).
Explore these bioinformatics educational opportunities and resources and share your feedback and ideas to improve this blog. Also, if you know of additional resources that we can add to the list, please send all correspondence to Maze Ndukum at ndukummaze@wustl.edu or askbecker@wustl.edu.